The Annotate SDK revolves around three major concepts: Projects, Datasets, and Ontologies. Each of these concepts is linked to a class that allows you to interact with them using the Annotate SDK. For example, the Project has an associated Project class and the Dataset has an associated Dataset class.
Note
Install the Annotate SDK by following the Installation guide or go to the Authentication page to get started with the Encord Annotate SDK.
Users
Users are people authenticated to interact with the Encord platform. Users can access resources and capabilities that are also available on the platform. For a Web app specific description of capabilities, go to the Annotate page.
Tip
For collaborative teams using our SDK, we recommend creating shared service accounts and creating SSH keys for those shared accounts. For example, to have several people create ontologies, datasets, and projects programmatically, create an email account for use with Encord (for example, [email protected]) and generate an SSH for that email account.
Datasets
Datasets are the space where the data itself lives. Datasets can be reused across multiple projects and contain no labels or annotations themselves. For a Web app specific introduction, go to the Datasets page.
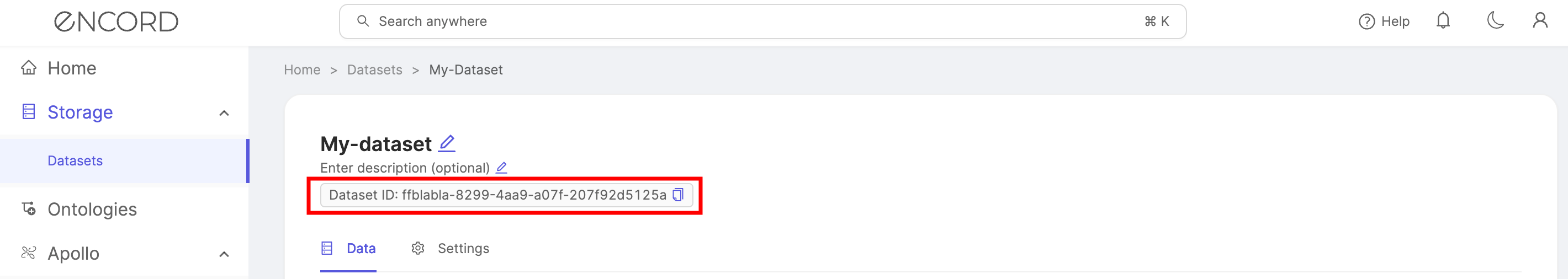
Each dataset is identified using a unique "<dataset_hash>" - a unique ID that can be found within the dataset's Settings, or the URL in the Encord platform when the dataset is being viewed, as shown below.
Data units
Datasets are composed of data units - packages of data that constitute a single annotation task. For example, a video, a single image, an image group, or a DICOM series. Data units are identified using their `"<data_unit_hash>", which can be found in the URL when the data unit is open in your browser, as shown below.
app.encord.com/label_editor/<data_unit_hash>&<project_hash>/0/0
Note
The
<data_unit_hash>and the<project_hash>are both shown in the URl when a data unit is open in the browser. They are separated by the '&' symbol, as shown above.
Projects
The space where labels, annotations, and reviews live. Projects can be linked to multiple datasets. For a Web-app specific introduction, go to the Dataset page.

